Technology Editor, Nature
This manuscript (permalink) was automatically generated from jperkel/mymanuscript@b2f0193 on March 31, 2020.
To provide a working example for an article on collaborative science writing platforms, we created this demonstration document. The article was published 31 March 2020 in Nature [1]. We encourage readers to explore and contribute to this document, as detailed in the Methods section below.
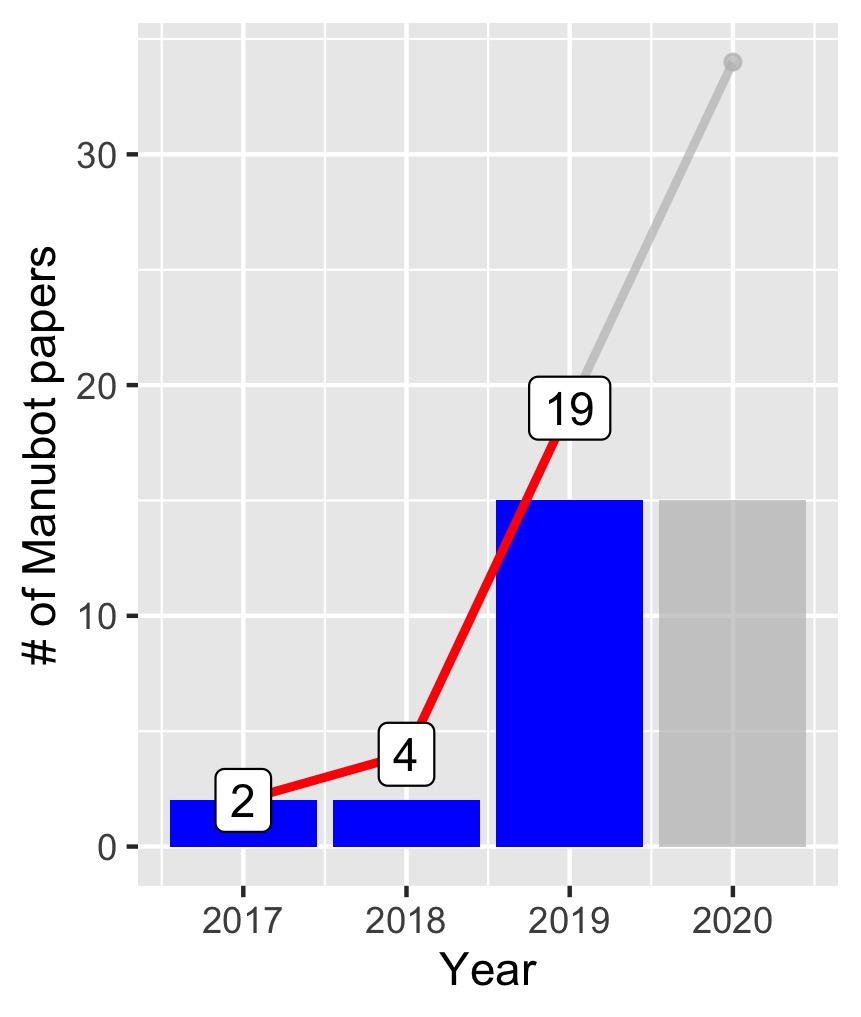
Manubot is a tool for collaboratively authoring and editing scientific manuscripts. It marries the scientific writing process with the workflow of open-source software development, much of which occurs on GitHub, and more than 30 articles have been published online using the tool (Figure 1).
Authors and collaborators can contribute to a project either by editing the document files on the GitHub web site or by cloning the repository to their own computer and editing the text there. In either case, changes are submitted in the form of a GitHub ‘pull request’, which allows the project maintainers to review the proposed edits.
Casey Greene – whose lab first developed Manubot to manage a review of deep learning – added the following text to this document via pull request:
Let’s add a citation by persistent identifier. This is my favorite feature of Manubot [2]. It will also give you a chance to review a pull request.
Greene’s pull request added a reference to the original Manubot paper using just its DOI – a feature called ‘cite-by-identifier’. Citations can also be added using arXiv [3] or PubMed IDs [4], URLs [5], and other identifiers. Here we’ll add a second reference by DOI. [6]. Note the tooltip that appears if you mouse over the inline references, which provides a direct link to the PubMed record, among other information. As this particular reference is actually cited twice in this article, you should also see navigation arrows that allow you to jump to each location.
Manubot builds documents from text files written in the simple formatting language called Markdown, in which, for example, bold text is indicated by double asterisks and italic text is indicated by single asterisks. You can add superscripts (x2) and subscripts (H2O), as well as strikethrough text, hyperlinks, and more.
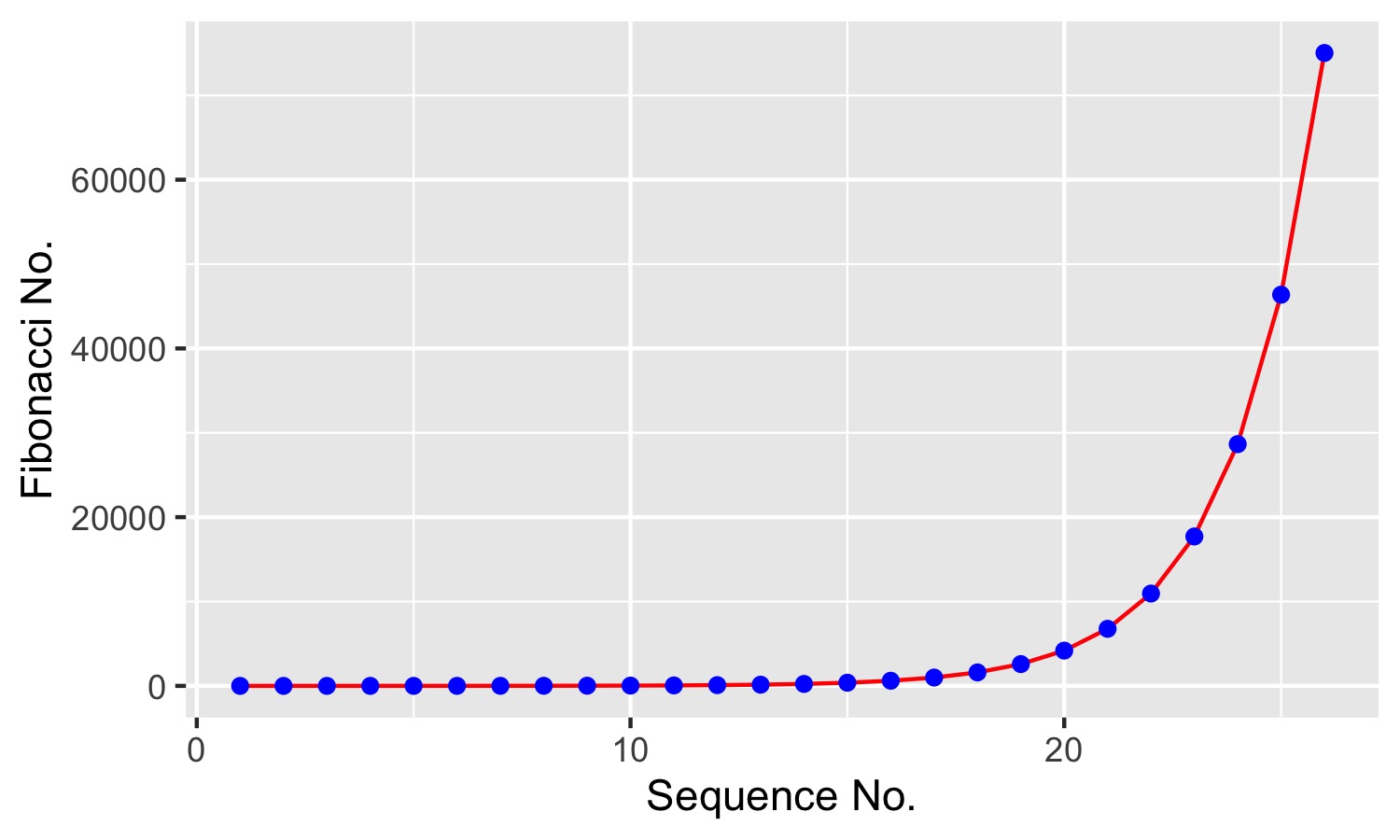
Here we’ll demonstrate the process of inserting data tables and figures, using the Fibonacci sequence as an example.
The Fibonacci sequence is a numeric sequence in which each number is the sum of the previous two numbers (0, 1, 1, 2, 3, 5, …). Expressed mathematically (using \(\LaTeX\)):
\[F_n = F_{n-1} + F_{n-2}\]
We can calculate that sequence using the programming language R, and we can show that code here in Markdown:
# calculate the first `n` Fibonacci numbers
fibonacci <- function(n) {
stopifnot(n > 2)
ar <- rep(0, n)
ar[1] <- f1 <- 0
ar[2] <- f2 <- 1
for (i in 3:n) {
f3 <- f1 + f2
ar[i] <- f3
f1 <- f2
f2 <- f3
}
return (ar)
}
f <- fibonacci(26)Manubot treats figures and tables the same as citations, and numbers them automatically by order of appearance. Authors can reference those objects in their text using identifiers, which take the form of an ‘at-symbol’ followed by object-type:identifier. DOIs are cited using doi:DOI – doi:10.1038/d41586-019-03632-y, for instance, while tables are identified by tbl:table-identifier [6]. The first 26 Fibonacci numbers are shown in Table 1.
| No. | Fib. No. | No. | Fib. No. |
|---|---|---|---|
| 1 | 0 | 14 | 233 |
| 2 | 1 | 15 | 377 |
| 3 | 1 | 16 | 610 |
| 4 | 2 | 17 | 987 |
| 5 | 3 | 18 | 1597 |
| 6 | 5 | 19 | 2584 |
| 7 | 8 | 20 | 4181 |
| 8 | 13 | 21 | 6765 |
| 9 | 21 | 22 | 10946 |
| 10 | 34 | 23 | 17711 |
| 11 | 55 | 24 | 28657 |
| 12 | 89 | 25 | 46368 |
| 13 | 144 | 26 | 75025 |
Figures are indicated by fig:figure-identifier. For instance, a graphical representation of the data in Table 1 is shown in Figure 2. (Note the tooltip that appears if you hover over the preceding figure number – it is a thumbnail of the figure itself.)
To create or contribute to a Manubot project, you will need to be logged into GitHub. If you do not have an account, create one.
rootstock repository and following the instructions in the repo’s setup.md file.content directory.content subdirectory.00.front-matter.md, 01.abstract.md, …). In the gray bar above the document text is a toolbar; select the pencil icon to edit the file. Make your suggested changes.This tutorial video demonstrates the process.
The following code was used to generate the Manubot data figure shown in Figure 1.
library(tidyverse)
library(gghighlight)
# data from https://manubot.org/catalog/ as of 31 Mar 2020
df <- read.csv(textConnection(
"Year,N
2017,2
2018,2
2019,15
2020,15"
))
# compute cumulative total for each year
df["cumsum"] <- cumsum(df$N)
p <- ggplot (df) +
geom_col(aes(x=Year, y=N), fill="blue") +
geom_line(aes(x=Year, y=cumsum), color = "red", size = 1) +
geom_point(aes(x=Year,y=cumsum)) +
gghighlight(Year < 2020, use_direct_label = FALSE) +
geom_label(aes(Year, cumsum, label=cumsum)) +
ylab("# of Manubot papers")
print(p)
ggsave('~/Downloads/manubot.jpg', width = 2.87, height = 3.39, units = "in")1. Synchronized editing: the future of collaborative writing
Jeffrey M. Perkel
Nature (2020-03-31) https://doi.org/ggqk8s
DOI: 10.1038/d41586-020-00916-6
2. Open collaborative writing with Manubot
Daniel S. Himmelstein, Vincent Rubinetti, David R. Slochower, Dongbo Hu, Venkat S. Malladi, Casey S. Greene, Anthony Gitter
PLOS Computational Biology (2019-06-24) https://doi.org/c7np
DOI: 10.1371/journal.pcbi.1007128 · PMID: 31233491 · PMCID: PMC6611653
3. First M87 Event Horizon Telescope Results. I. The Shadow of the Supermassive Black Hole
The Event Horizon Telescope Collaboration
arXiv (2019-07-08) https://arxiv.org/abs/1906.11238
DOI: 10.3847/2041-8213/ab0ec7
4. A new coronavirus associated with human respiratory disease in China.
Fan Wu, Su Zhao, Bin Yu, Yan-Mei Chen, Wen Wang, Zhi-Gang Song, Yi Hu, Zhao-Wu Tao, Jun-Hua Tian, Yuan-Yuan Pei, … Yong-Zhen Zhang
Nature (2020-02-03) https://www.ncbi.nlm.nih.gov/pubmed/32015508
DOI: 10.1038/s41586-020-2008-3 · PMID: 32015508 · PMCID: PMC7094943
5. CRISPR treatment inserted directly into the body for first time
Heidi Ledford
Nature (2020-03-05) https://www.nature.com/articles/d41586-020-00655-8
DOI: 10.1038/d41586-020-00655-8
6. The microscope makers putting ever-larger biological samples under the spotlight
Jeffrey M. Perkel
Nature (2019-11-26) https://doi.org/ggm9g5
DOI: 10.1038/d41586-019-03632-y · PMID: 31772373